1/8ページ
ダウンロード(1.2Mb)
Application Note 26「Octet potency assay: development, qualification and validation strategies」
ホワイトペーパー
このカタログについて
| ドキュメント名 | Application Note 26「Octet potency assay: development, qualification and validation strategies」 |
|---|---|
| ドキュメント種別 | ホワイトペーパー |
| ファイルサイズ | 1.2Mb |
| 取り扱い企業 | ザルトリウス・ジャパン株式会社 (この企業の取り扱いカタログ一覧) |
この企業の関連カタログ

このカタログの内容
Page1
APPLICATION NOTE 26
Octet potency assay: development, qualification
and validation strategies
Carson Cameron, Brendan Peacor, Nathan Oien, Andrew Cheeseman, and Jimmy Smedley, KBI Biopharma, Durham, NC
John Laughlin, and David O. Apiyo, ForteBio, Fremont, CA
Introduction
Kinetic analysis of biomolecular interactions is critical during development and validation of a method for evaluating the
drug discovery and development. The affinity of an interaction binding of an Fc gamma receptor III molecule to the widely
directly affects the dose required for a biopharmaceutical to be characterized NISTmAb.
effective. Real-time data on affinity and the kinetics of binding
can provide useful information at every stage of biopharma- Fc receptors are widely distributed cell-surface proteins that act
ceutical reagent development. Moreover, understanding the as communication points between effector antibodies and their
mechanism of binding can provide insights into the desirability biological implements. There are three classes of Fc recep-
of a drug candidate during development, including implications tors including Fc-gamma receptor I (CD64) that is responsible
for the drug’s stability upon complex formation with its binding for phagocytosis and the activation of monocytic cell lines,
target. Binding kinetics assays and specifically affinity constant Fc-gamma receptor II (CD32) that is mainly responsible for anti-
(K body-dependent cellular phagocytosis and Fc-gamma receptor
D) analysis are increasingly being used for biological products
lot release. Regulatory requirements necessitate that such III (CD16), which is responsible for antibody-dependent cellular
products be QC tested using methods that are robust and have cytoxicity (ADCC). These receptor molecules bind to antibodies
been appropriately developed, qualified and validated under through their Fc region and impart different activities. Glycosyla-
GMP conditions. tion and other modifications to the Fc region of an antibody can
affect Fc gamma receptor binding hence these receptor mole-
In this application note, we discuss the strategies for the cules are a good tool for evaluating antibody drug efficacy and
development and validation of a potency assay using Octet® for antibody product lot release assessment. In this application
systems. We have highlighted the Octet system’s ease-of- note, we use affinity constants (KD) as the reportable parameter
use and fast time to results by showcasing strategies for the to determine Percent Relative Potency to a reference lot.
1.0
Incident BLI signal
white processing 0.8
light
0.6
Biocompatible
surface 0.4
Bound
molecule 0.2
Unbound molecules
have no effect
Wavelength (nm)
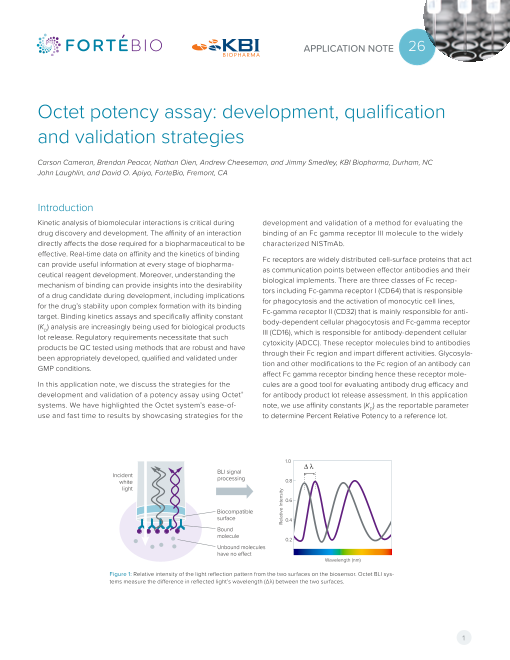
Figure 1: Relative intensity of the light reflection pattern from the two surfaces on the biosensor. Octet BLI sys-
tems measure the difference in reflected light’s wavelength (Δλ) between the two surfaces.
1
Relative Intensity
Page2
Bio-Layer Interferometry Three key input variables: temperature, shake speed and ligand
loading density, were identified to be critical to the performance
The Octet platform utilizes a Dip and Read™ format in combina- of the potency assay and were evaluated in a mini-design of
tion with Bio-Layer Interferometry (BLI) to monitor the interac- experiment (DOE) prior to establishing the method. Each vari-
tions between biological molecules. BLI is an optical technique able was examined at three or four levels. To establish assay
where white light incident to a reflective biosensor surface performance at these conditions, the analyte concentration,
immobilized with a ligand results in changing interference pat- sample matrix and assay step run times were maintained at con-
terns of the reflected white light detected upon the interaction stant values. A control condition was set at the ForteBio default
between the ligand and the analyte which is kept in solution. conditions for ligand binding assay (kinetics) with the shaking
Binding events between the ligand and the analyte result in an speed at 1000 RPM and temperature at 30°C for these studies.
increase in optical thickness on the tip of the biosensor that can
be measured as a wavelength shift from the reference surface, Optimal assay behavior can be split into four distinct parts for
and is a proportional measure of the change in thickness of the kinetic determination assays:
biological layer (Figure 1). 1 The baseline(s) must be flat and absent of upward or down-
ward drift (Figure 2, 0–60 seconds).
Materials and reagents
2 The loading density should be adequate to ensure sufficient
Materials and reagents for the studies: protein is loaded on the biosensor, but not too high as to
cause steric hindrance of the subsequent binding event.
Material/reagent Vendor Catalog #
Multiple concentrations of the loading protein (Figure 2) are
Ni-NTA biosensors ForteBio 18-5102 typically evaluated, often with a following association step
FcγRIIIa R&D Systems 4325-FC-050 although this is not mandatory. In general, low loading densi-
ty is recommended when using Ni-NTA biosensors.
NISTmAb National Institute RM8671
of Standards and 3 The association step should show a concentration-de-
Technology (NIST)
pendent signal over at least 1.5 orders of magnitude (in
96-well plates Griener Bio-One 655209 concentration) as seen in Figure 2 (0 to 300 seconds). This
384-well plates Griener Bio-One 781209 step should also not show binding heterogeneity unless it is
known that there is a 2:1 binding event occurring as indicat-
Octet HTX system ForteBio NA
ed by a sigmoidal curve (similar to Figure 2) followed by a
Diluent (1X PBS, KBI NA linear increase in signal rather than a plateau.
0.1%Tween 20, 0.2% BSA) (prepared day of use)
4 The dissociation step should show at least a 5 % drop in
binding signal (Figure 4, 300 to 900 seconds).
Method development
Determination of analyte characteristics, including potency, DATA ACQUISITION SETUP
can be affected by the test method used. Factors such as the For most of the kinetics experiments, a 384-well plate was used
amount of FcγRIIIa immobilized on the biosensor for the detec- to enable high-throughput development (example shown in
tion of the analyte, the temperature and shaking speed of the Figure 3). Additional buffer and loading wells were added as
reaction, the sample matrix, and the equilibration time allowed needed. Control points were performed at the beginning of a
for binding can affect the binding behavior. Each of these run prior to setting the shake speed or temperature to ensure
factors can also impact critical assay attributes such as accu- no mechanical effects (i.e. degradation due to increased tem-
racy. The Octet platform is highly suited for a fast evaluation perature) on the control sample.
of the interactions between these potential key assay inputs
and allows for relatively high-throughput method development.
2
Page3
BIOSENSOR SELECTION 2.5
Ni-NTA biosensors were selected for this assay for robustness,
ease of use, and to take advantage of the commercial availabil-
2.0
ity of poly-histidine tagged (HIS-tag) receptors. FcγRIIIa is avail-
able commercially with many different purification conjugates.
Using FcγRIIIa with a histidine tag at the C-terminus ensured 1.5
the optimal orientation of the protein binding to the biosensor
and provided the most distance between the biolayer of the
biosensor and the binding site to prevent hindrance of FcγRIIIa 1.0
binding to NISTmAb.
A1
0.5
LIGAND DENSITY ASSESSMENT B1
C1
D1
Proteins such as Fc receptors typically provide optimal load-
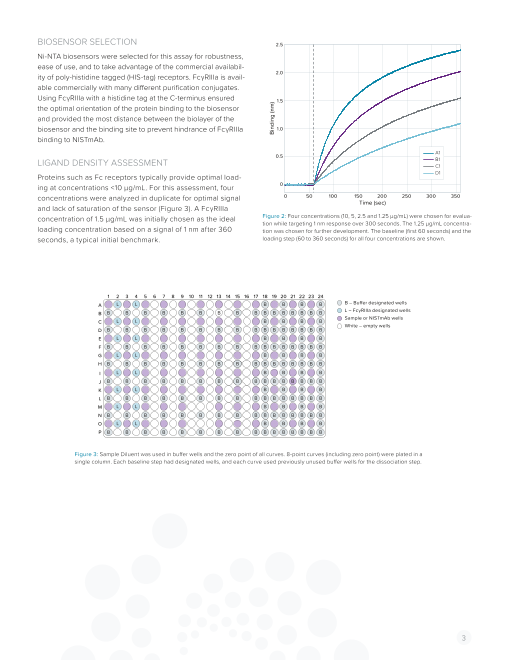
ing at concentrations <10 µg/mL. For this assessment, four 0
concentrations were analyzed in duplicate for optimal signal 0 50 100 150 200 250 300 350
Time (sec)
and lack of saturation of the sensor (Figure 3). A FcγRIIIa
concentration of 1.5 µg/mL was initially chosen as the ideal Figure 2: Four concentrations (10, 5, 2.5 and 1.25 µg/mL) were chosen for evalua-
tion while targeting 1 nm response over 300 seconds. The 1.25 µg/mL concentra-
loading concentration based on a signal of 1 nm after 360 tion was chosen for further development. The baseline (first 60 seconds) and the
seconds, a typical initial benchmark. loading step (60 to 360 seconds) for all four concentrations are shown.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24
A L L B B B B B – Buer designated wells
B B B B B B B B B B B B B B B B B L – FcγRIIIa designated wells
C L L B B B B Sample or NISTmAb wells
White – empty wells
D B B B B B B B B B B B B B B B B
E L L B B B B
F B B B B B B B B B B B B B B B B
G L L B B B B
H B B B B B B B B B B B B B B B B
I L L B B B B
J B B B B B B B B B B B B B B B B
K L L B B B B
L B B B B B B B B B B B B B B B B
M L L B B B B
N B B B B B B B B B B B B B B B B
O L L B B B B
P B B B B B B B B B B B B B B B B
Figure 3: Sample Diluent was used in buffer wells and the zero point of all curves. 8-point curves (including zero point) were plated in a
single column. Each baseline step had designated wells, and each curve used previously unused buffer wells for the dissociation step.
3
Binding (nm)
Page4
A B
0
0.30
-0.10
0.20
-0.20
0.10
-0.30
0
0 200 400 600 800 0 200 400 600 800
Time (sec) Time (sec)
C D
1.5 1.5
1 1
0.5 0.5
0 0
-0.5 ND40 -0.5
ND41
-1 -1 ND40
ND51
-1.5 -1.5
0 5 10 15 20 25 30 35 40 45 0 5 10 15 20 25 30 35 40 45
Figure 4: A) Inverted signal of NISTmAb associating to a FcγRIIIa bound to the bisosensor tip (loading of FcγRIIIa not shown). B) Flipped data from A using the “Flip Data”
feature on Octet Data Analysis software. C) Typical wavelength shift (left to right) from a small change in optical thickness (ND = Optical Thickness). D) Wavelength shift
from a large change in optical thickness (right to left) resulting in an inverted signal.
ANTIBODY BINDING Step Step type Time (s) Shaker speed (RPM)
NISTmAb concentration scouting was performed beginning with 1 Baseline 60 1000
a range of 500 to 1.56 µg/mL. Curve shape, RMax, Chi2, R2, Glob-
2 Loading 300 1000
al Fit vs Local Fit, and Steady State were all considered when
determining the optimal antibody binding. These attributes were 3 Baseline 2 120 1000
also evaluated when establishing data processing parameters. 4 Association 300 1000
The working range was determined to be 200 to 3.125 µg/mL
5 Dissociation 600 1000
based on acceptable assay performance. During the association
step, signal inversion occurred. Signal inversion is a less-known Table 1: Octet assay steps.
phenomena that arises when the optical thickness at the tip of
the biosensor experiences a large change (Figure 4).1 This is PRE-QUALIFICATION/VALIDATION ASSESSMENT
usually attributed to large molecules or complexes binding to a
biosensor and is indicated as a decrease in signal. To verify that For NISTmAb binding to FcγRIIIa, a short screening assay was
binding is occurring and not dissociation, the decrease in signal performed as described above to determine the optimal Fcγ-
should be concentration-dependent and often a following step RIIIa loading concentration and NISTmAb concentration range.
should be included, such as a dissociation step. The final assay A DOE was then planned (Table 2) to determine the optimal
steps are shown in Table 1 and were used for the theoretical loading concentration and if the platform conditions (30°C and
pre-qualification/validation assessment. 1000 RPM) were suitable. A control preparation was performed
4
Binding (nm)
Binding (nm)
Page5
at these platform conditions (including 1.5 µg/mL loading) to equivalent to reference. The results of an Octet %relative poten-
observe day-to-day repeatability and to calculate %relative cy method qualification generally allow criteria to be set for: R2,
potency. The DOE approach, coupled with the fast assay time X2, maximum response signal, minimum response signal, and a
of the Octet platform allows for the method parameters to be range of % Relative Potency (potency comparison to reference).
scouted in minimal time. Typically, the results from running a qualified method (during
development or stability experiments, etc.) in conjunction with
Parameter Range Number of points the process (purification, culture, etc.) can provide the data to
Temperature 28–35°C 4 set criteria for a validation protocol. Validation of a method com-
monly repeats the studies, (linearity, accuracy, etc.) performed
Loading concentration 0.75–3.0 µg/mL 4 in the qualification with tighter passing criteria and includes
Shake speed 800–1200 RPM 3 significant ruggedness and robustness studies.
Table 2: Pre-qualification/validation development DOE.
METHOD VALIDATION
The results were analyzed using statistical analysis software Method Validations are completed to ensure an analytical
which showed the optimal conditions for this assay were a 1.1 method is suitable for its intended purpose. This provides
(±0.1) µg/mL loading concentration for FcγRIIIa, a 1000 (±100) RPM an assurance of reliability for routine testing in GMP envi-
plate shake speed, and a 30°C (29.5-31.5°C) assay temperature. ronment. Validation involves comprehensive protocol-driven
experiments that evaluate and document the performance of
CRITICAL PROCESS PARAMETER ASSESSMENT an assay. As this method was being established as a potency
The pre-qualification DOE also provided the data required assay, linearity, specificity, accuracy, precision, range, robust-
to assess specificity, precision/repeatability, and the working ness, and ruggedness were evaluated as recommended by
range of the assay. A diluent blank was performed as part ICH Guideline Q2 (R1)3 “Validation of Analytical Procedures:
of each NISTmAb curve. These blanks all demonstrated no Text and Methodology.”
matrix interference, indicating specificity of the assay. Due to Linearity is the expected relationship between known poten-
the nature of the DOE, evaluating precision required assessing cies of samples and their measured values using a range of
the data points from center points of the DOE. The average 50% to 150% of the nominal relative potency, but treating them
%relative potency was 91% with a %RSD of 7%, suggesting good as 100%. Five levels were tested over the 50% to 150% range
precision of the assay. Further, all points in the DOE showed R2 including 100%. The R2 values of the resulting curves were all
≥0.97, suggesting the working range of the assay (200 µg/mL to ≥0.95, indicating good linearity.
3.125 µg/mL) is suitable for qualification.
Accuracy is the degree of closeness to the expected value and
The hydration of the biosensors was also evaluated. The base- was determined using results obtained from the linearity studies
line signal immediately after biosensor hydration of 10, 15, and by calculating the percent recovery for each linearity level. For
20 minutes was comparable, demonstrating that a 10- minute example; a %Relative Potency of 46% at the 50% linearity level
hydration time was suitable for the final method. returns a 92% recovery. The average %recovery was calculated
to be 97% with a range of 85% to 118% recovery. These results
METHOD QUALIFICATION showed the method was accurate.
Method Qualification, while not always required, can be a useful Precision is the variability in the data from replicate determi-
tool in early phases of drug development and provide critical nations under normal assay conditions. Repeatability of the
data leading up to a validation. In general, the qualification of method was assessed by testing multiple preparations at the
a potency method involves evaluating linearity, specificity, ac- nominal load. The average relative potency was 101% with a
curacy, precision, and range. Method Qualification also serves %RSD of 6%. Intermediate precision of the method was as-
to set system suitability criteria for the assay as well as sample sessed using a second analyst to test multiple preparations at
acceptance criteria for release testing and/or stability samples. the nominal load. The average relative potency between two
For instance, the results from the accuracy calculations may analysts was 101% with a %RSD of 8%. These results were within
allow for a criterion of 70% to 130% relative potency for test arti- the expected limit.
cles. When test samples meet this criterion, they are considered
5
Page6
The range of the method is demonstrated when precision, ac- %Relative
curacy and linearity of the method show suitable performance. Run Biosensor FcyR3A Analyst KD (nM) potency
Suitable performance was demonstrated spanning the working 1 1 2 1 25 100%
range of 50% to 150% of the nominal potency. This correspond-
ed to 100 µg/mL to 300 µg/mL for the highest concentration of 2 1 2 3 21 93%
the dose-response curve. 3 5 1 1 31 82%
Specificity of the method was verified by testing a buffer blank 4 1 2 2 30 109%
and a generic non-human antibody, both diluted in the same 5 1 1 2 28 103%
scheme as NISTmAb. The %relative potency of the blank and
6 2 1 1 23 112%
generic antibody were determined to be not-comparable to
NISTmAb and specificity of the method was confirmed. 7 3 2 2 25 91%
Robustness of this assay was evaluated by testing the work- 8 4 2 3 24 80%
ing range of the parameters generated by the results of the 9 5 2 1 21 119%
development DOE. This involved making small but deliberate 10 3 1 1 22 114%
changes to the assay loading concentration, shake speed, and
11 5 1 2 30 110%
temperature. These changes in methodology returned results
within 70% to 130% proving the method is robust. 12 3 2 3 22 88%
Ruggedness of this assay was tested by evaluating normal test 13 5 2 2 26 96%
conditions that may vary over time. To test ruggedness of the 14 3 2 1 20 127%
assay, a DOE was performed on the parameters with the most 15 5 1 2 28 113%
risk for variance. This included biosensor lot, FcγRIIIa lot, and
analyst to analyst variability (Tables 3 and 4). 16 1 1 2 22 86%
17 4 2 1 23 108%
The results of the DOE were analyzed by performing a Fit
Least Squares analysis. The results of this analysis are shown 18 3 1 2 27 105%
in Figure 5. The Effect Summary Table showed no statistically 19 5 2 1 24 104%
significant interactions (i.e. all ρ values were greater than 0.05). 20 1 1 1 19 109%
The Prediction Profile and Interaction Profiler showed no clear
substantial trends between different variables. The effects of 21 1 2 2 25 98%
this DOE prove the method is rugged. 22 1 1 3 18 105%
23 4 2 3 18 109%
Parameter # of Points
24 3 1 2 30 115%
Biosensor lots: 5 25 3 2 2 22 86%
FcγR3A lots: 2 26 1 2 1 23 109%
Analysts: 3 27 5 1 1 31 82%
Table 3: Ruggedness DOE. 28 5 2 2 25 97%
29 1 1 1 25 85%
30 3 2 1 33 76%
Table 4: A 30 run DOE showing the various combination of parameters tested.
6
Page7
A Effect summary table
Source LogWorth PValue
Biosensor*FcyRIIIa 0.677 0.21029
Biosensor*Analyst 0.629 0.23475
Biosensor 0.611 0.24497
Biosensor*FcyRIIIa*Analyst 0.551 0.28098
FcyRIIIa 0.216 0.60810
FcyRIIIa*Analyst 0.171 0.67448
Analyst 0.079 0.83416
B Prediction profiler
130
120
110
100
90
80
70
1 2 3 4 5 1 2 1 2 3
1 1 1
Biosensor FcyRIIIa Analyst
C Interaction profiler
130
120 3 3
110 2 2
1 1
100 Biosensor
90
80 5 5
4 4
70
130
120
110 2
1
100 FcyRIIIa
90 2
1
80
70
130 3
120
110 2
100 Analyst
1
90 2 3
1
80
70
1 2 3 4 5 1 2 1 2 3
Figure 5: A) Effect screen of multiple parameters showing no significant interactions. LogWorth = -Log (p-Value). B) Pre-
diction Profiler showing results are not able to predict future trends in data. C) Interaction Profiler showing the interactions
between two variables have no predictable effect on %Relative Potency.
7
%Relative Potency
97
%Relative Potency %Relative Potency %Relative Potency [75.0558, 118.944]
Biosensor FcyRIIIa Analyst
Page8
Results Parameter Reportable result Result
A %Relative Potency method for FcgammaR3 has been devel- Linearity R2 of triplicate preps R2 ≥ 0.95
oped and analyzed in a representative method validation. For Specificity Diluent and non-specific Not comparable
this validation exercise, the representative raw data can be mAb comparable to
seen in Figure 6 and analyzed results in Table 5. The results reference
show that this method is linear, specific, accurate, precise, ro- Accuracy %Recovery of linearity 85% to 118% recovery
bust, and rugged over a specific range in accordance with ICH preparations
Guidelines Q2 (R1).3 Repeatability Average %relative Average = 101%,
potency and %RSD %RSD = 6%
Octet systems in GxP laboratories Intermediate Average %relative Average = 101%,
precision potency and %RSD of %RSD = 8%
The use of Octet systems in GxP laboratories is constantly Analyst I and Analyst II
expanding. KBI Biopharma has successfully developed 30+
Range Method range 50% to 150% for
methods on the Octet platform used for titer, potency, kinetics, highest concentration
and identity testing. Many of these methods are being used
to support Manufacturing, Drug Substance or Drug Product Robustness %Relative potency at 70% to 130% Relative
modified conditions Potency
Release testing, and Long-term Stability testing in a GxP envi-
ronment. While the assay and sample acceptance criteria are Ruggedness Results of DOE No significance from
parameters or inter-
dependent on the method variability as well as the process actions
variability, these methods generally exhibit ≤10 %RSD between
replicates over long term testing. Table 5: Results of the validation exercise.
Conclusion
Functional biological activity is a critical quality attribute (CQA)
essential to verifying the potency of a drug molecule.2 Potency
0.30 assays can be used throughout the development process in
comparability and formulation studies, and are required for
release of every lot of therapeutic protein. The Octet plat-
0.20 form offers a fast, accurate, and robust solution for measuring
potency of a drug molecule. Here we have described consid-
erations for the development of a %relative potency method
capable of early-phase comparability studies and subsequent
0.10 method validation for lot release. With the speed of the Octet
HTX system, we could rapidly achieve Design of Experiment
results which led to development, optimization, and potential
validation practices.
0
0 200 400 600 800 References
Time (sec)
1 Application and Comparison of Algorithms for the Evaluation of
Figure 6: Replicate binding curves (n=6) of NISTmAb binding to FcγRIIIa. Interferograms, G. Kraus and G. Gauglitz Fresenious’ J. Anal. Chem.,
349: 399–402, 1994.
2 ICH Guideline Q6B, “Specifications: Test Procedures and Acceptance
Criteria for Biotechnological/Biological Products.”
3 ICH Guideline Q2R1, “Validation of Analytical Procedures: Text and
Methodology.”
ForteBio ForteBio Analytics (Shanghai) Co., Ltd. Molecular Devices (UK) Ltd. Molecular Devices (Germany) GmbH
47661 Fremont Boulevard No. 88 Shang Ke Road 660-665 Eskdale Bismarckring 39
Fremont, CA 94538 Zhangjiang Hi-tech Park Winnersh Triangle 88400 Biberach an der Riss
888.OCTET-75 or 650.322.1360 Shanghai, China 201210 Wokingham, Berkshire Germany
www.fortebio.com fortebio.info@moldev.com salesops.china@moldev.com RG41 5TS, United Kingdom + 00800 665 32860
+44 118 944 8000
uk@moldev.com
©2019 Molecular Devices, LLC. All trademarks used herein are the property of Molecular Devices, LLC. Specifications subject to change without
notice. Patents: www.moleculardevices.com/product patents. FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES.
41-0298-AN Rev B
Binding (nm)